Improved Pogona Genome Assembly
It is with great pleasure that we are able to announce the submission of a telomere to telomere genome assembly for a female ZW individual of the Australian bearded dragon Pogona vitticeps. There is a genome assembly from the BGI team that is also worth a look -- it is for a male ZZ individual. The two papers are to appear concurrently in the journal GigaScience.
The central bearded dragon is widely distributed in central eastern Australia and adapts readily to captivity. Among other attributes, it is distinctive because it undergoes sex reversal from ZZ genotypic males to phenotypic females at high incubation temperatures.
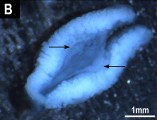
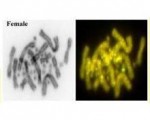
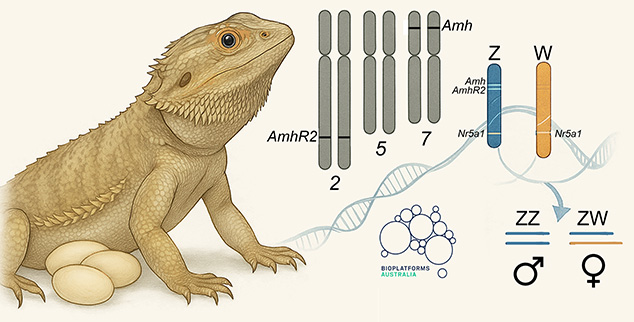
Genome assembly length is 1.75 Gbp with a scaffold N50 of 266.2 Mbp, N90 of 28.1 Mbp, 26 gaps and 42.2% GC content. Most (99.6%) of the reference assembly is scaffolded into 6 macrochromosomes and 10 microchromosomes, including the Z and W microchromosomes, corresponding to the karyotype.
The genome assembly exceeds standard recommended by the Earth Biogenome Project: 0.003% collapsed sequence, 0.03% false expansions, 99.8% k-mer completeness, 97.9% complete single copy BUSCO genes and an average of 93.5% of transcriptome data mappable back to the genome assembly. The mitochondrial genome (16,731 bp) and the model rDNA repeat unit (length 9.5 Kbp) were assembled. All major scaffolds were chromosome length, with terminal telomeres. The assembly was remarkably gap free (26 gaps in all) and had a very low error rate (1 error in 146 Kbp).
Male vertebrate sex genes Amh and Amhr2 were discovered as copies in the small non-recombining region of the Z chromosome, absent from the W chromosome.
This, coupled with the prior discovery of differential Z and W transcriptional isoform composition arising from pseudoautosomal sex gene Nr5a1, suggests that complex interactions between these genes, their autosomal copies and their resultant transcription factors and intermediaries, determines sex in the bearded dragon. The more you look, the more you see.
There are a few nuances in this assembly. First of all, we could not identify the centromeric region on the basis of distinct repetitive satellite units, as we did in the Bassiana(=Acritoscincus) duperreyi genome assembly. There also no signature in the traces of read depth or GC content which enabled us to locate the centromeres as we did in the Bassiana duperreyi genome assembly. This was very frustrating. Instead we found a signature in terms of interchromosomal HiC contact in the vicinity of putative centromeres, and a co-located precipitous drop in the heterozygosity rate. These putative centromeric regions coincided with the centromeres of the physical images of the Pogona chromosomes. Clearly, the characterization of centromeric sequence of Pogona vitticeps in a comparative study with other squamates would make an interesting study.
A second interesting thing, this time about the assembly process, was that mitochondrial sequence was absent from the PacBio HiFi data. We assume it was eliminated during the size selection step. As the assembly software hifasm uses PacBio HiFi for the core assembly, these mitochondrial sequences, although present in the ONT data, were not recovered from the ONT data during the combined assembly process. We also noted a drop in the read depth for the PacBio HiFi and Illumina data for exceptionally small microchromosomes that is not observed in the ONT data. This suggests a systematic bias in the data from sequence-by-synthesis platforms for small elements and high GC content. Similar issues seem to affect the assembly of ultrasmall microchromosomes in birds (Mathers, 2025). This will all be of considerable interest to assembly affecionados.
The central bearded dragon is the focus of many studies now, nationally and internationally. This high-quality assembly will serve as a resource to enable and accelerate research into the unusual reproductive attributes of this species and for comparative studies across the Agamidae and reptiles more generally. We thank Bioplatforms Australia and their AusARG initiative for making this happen.
Pull out these two papers for a read of the primary work:
-
Guo, Q., Pan, Y., Dai, W., Guo, F., Zeng, T., Chen, W., Mi, Y., Zhang, Y., Shi, S., Jiang, W., Cai, H., Wu, B., Zhou, Y., Wang, Y., Yang, C., Shi, X., Yan, X., Chen, J., Cai, C., Yang, J., Xu, X., Gu, Y., Dong, Li, Q. 2025. A near-complete genome assembly of the bearded dragon Pogona vitticeps provides insights into the origin of Pogona sex chromosomes. GigaScience 14 [https://doi.org/10.1093/gigascience/giaf079].
- Patel, H.R., Alreja, K., Andre L.M. Martin-Reis. A.L., Chang, J.K., Chew, Z.A., Jung, H., Hammond, J.M., Deveson, I.W., Ruiz-Herrera, A., Marin-Gual, L., Holleley, C.E., Zhang, X., Lister, N.C., Whiteley, S., Xiong, L., Dissanayake, D.S.B., Waters, P.D., Georges, A.. 2025. A telomere to telomere phased genome assembly and annotation for the Australian central bearded dragon Pogona vitticeps. GigaScience 14 [https://doi.org/10.1093/gigascience/giaf079].
or go to some popular press articles or the GigaScience Editors Take for an easier read.